Find all vectors of recurrence states compatible with relationship graph
Source:R/helpers.R
compatible_rstrs.RdFinds all possible recurrence states for each recurrence compatible with the relationship graph, then takes the Cartesian product to get all vectors of recurrence states. For a recurrence to be a recrudescence, all edges between the recurrent infection and the immediately preceding infection must be clonal edges. For a recurrence to be a reinfection, all edges between the recurrent infection and any preceding infection must be stranger edges. All recurrences may possibly be relapses.
Arguments
- RG
Relationship graph; see
enumerate_RGs.- gs_per_ts
List of vectors of genotypes for each infection.
Value
Vector of strings (consisting of "C", "L", "I" for recrudescence, relapse, reinfection respectively) compatible with relationship graph.
Examples
MOIs <- c(2, 2, 1)
RG <- enumerate_RGs(MOIs, igraph = TRUE)[[175]]
#> Number of valid relationship graphs (RGs) is 250
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> =
#> |
gs_per_ts <- split(paste0("g", 1:sum(MOIs)), rep(1:length(MOIs), MOIs))
# 1st recurrence can't be recrudescence, 2nd recurrence can't be reinfection
plot_RG(RG, edge.curved = 0.2)
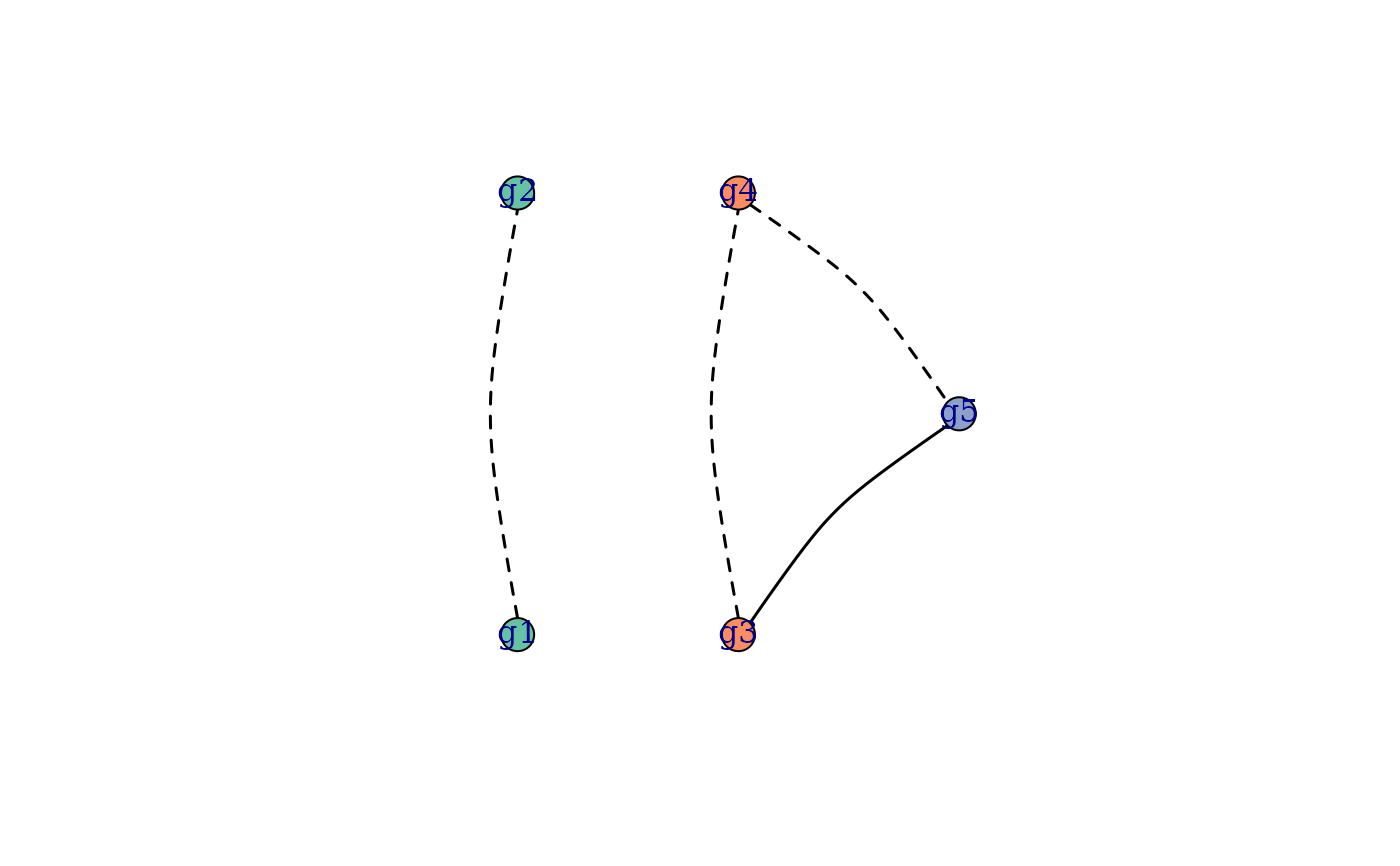 compatible_rstrs(RG, gs_per_ts) # "LL" "IL" "LC" "IC"
#> [1] "LL" "IL" "LC" "IC"
compatible_rstrs(RG, gs_per_ts) # "LL" "IL" "LC" "IC"
#> [1] "LL" "IL" "LC" "IC"